Group09 Report
| Overview | Proposal | Poster | Application | Report |
Contents
Introduction
Infectious diseases are caused by pathogenic microorganisms, such as bacteria, viruses, parasites or fungi; the diseases can be spread, directly or indirectly, from person to person, even from animals to humans. Zoonotic diseases are infectious diseases of animals that can cause disease when transmitted to humans.
The 21st century has already been marked by major epidemics. Old diseases - cholera, plague and yellow fever - have returned, and new ones have emerged - SARS, pandemic influenza, MERS, Ebola and Zika. These epidemics and their impact on global public health are quite remarkable.
Although disease patterns change constantly, communicable diseases remain the leading cause of mortality and morbidity in the least and less developed countries. Despite decades of economic growth and development in countries that belong to the World Health Organization (WHO) South-East Asia Region, most countries in this region still have a high burden of communicable diseases. This raises some urgent concerns. The first is that despite policies and interventions to prevent and control communicable diseases, most countries have failed to eradicate vaccine-preventable diseases. Second, sustainable financing to scale up interventions is lacking, especially for emerging and re-emerging diseases that can produce epidemics.
Objectives and Motivations
Diseases are prevalent no matter in which society, whilst, as the economy developing, healthcare becomes the major concern in daily life. Recently, there are still a lot of contagious diseases such as TB, malaria, cholera and meningitis, influenza A(H5N1) virus (avian flu), severe acute respiratory syndrome(SARS) and chikungunya reach high epidemic proportions in some countries, especially in developing countries. Thus, we want to apply visual analytics techniques to analyze historical records of seven contagious diseases: Smallpox, Rubella, Hepatitis, Measles, Polio, Mumps, Pertussis from US 1916-2010 and medical records of diseases and their corresponding symptoms. It can help us to find out patterns from these historical typical contagious diseases and apply to other diseases.
Scientific methods align with a huge amount of reliable researches always come out with the convincing and inspiring result. we intend to use a large-scale biomedical literature database to construct a symptom-based human disease network and investigate the connections with related diseases.
Nonetheless, this project also serves following purposes: 1) provide exploratory analysis of datasets; 2) aid domain experts seek for unexpected association among diseases as well as validate their research results; 3) bridge the gap between knowledge obtained by experts only as well as produced at the lab bench and its use at the clinical bedside; 4) non-specialists can gain straightforward and useful information (e.g. which symptom suspiciously causes a specific contagious disease) from the application.
Previous Work
Summary
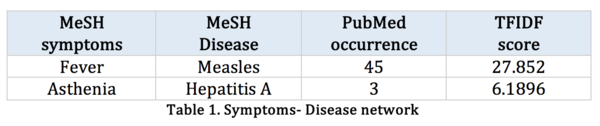
In human symptoms-disease [3], previous researchers used a large-scale biomedical literature database constructing a symptom-based human disease network and investigate the connection between clinical manifestations of diseases and their underlying molecular interactions. They demonstrated the similarity of two diseases correlates strongly with the number of shared symptoms.
Their research starts from crawling large-scale bibliographic records PubMed, they used its related Medical Subject Headings (MeSH) to extract symptom terms and disease terms from the bibliographies and applied text analysis techniques to generate co-occurrence and to calculate the TF-IDF score of each pair of symptoms and diseases. The dataset after their processing contains hundreds of diseases and thousands of symptoms.
Shortages
- It is very difficult to see the trend and spread area for those public users without any domain knowledge.
- Previous symptoms-disease network contains the majority of human diseases, the relationship among diseases and symptoms is obscured with such a large amount of data.
- Their data source is crawling medical reports from medical websites, as they have mentioned, the number of reports is less than the real incidences.
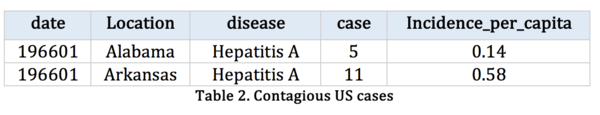
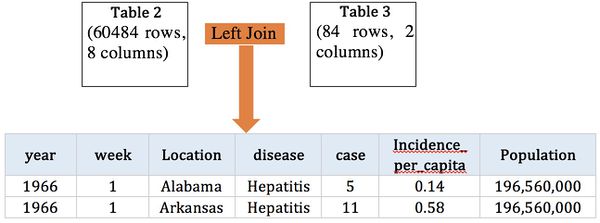
To improve, we obtain contagious disease records from Kaggle, which records the number of contagious incidence in US from 1916 to 2010.
Data process
Dataset Overview
|
Table view |
Description |
|
The symptoms-disease dataset is from Nature human symptoms-disease network (HSDN), which is the combination of the MeSH vocabulary and the PubMed literature. Filtering seven contagious diseases (same as below) for consistency purpose. | |
|
US contagious diseases from 1916-2010 | |
|
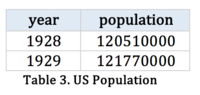
US population from 1916 -2010 |